

What do horse fetlocks, sperm tails, and chick embryos have in common? They're all being measured and studied in NCSA's virtual reality environment using software developed by the Biological Imaging Group at NCSA. The software system, named Crumbs, was developed in collaboration with the Beckman Institute and UIUC Department of Cell and Structural Biology. Crumbs marks a trail in 3D instrumental datasets.

As in the fable of Hansel and Gretel, Crumbs lets scientists explore paths or fibers in their datasets and drop bread crumbs, or markers, along the way to mark their trails. Paths can be measured and saved for quantitative analysis using NCSA's CAVE(tm) (Cave Automatic Virtual Environment). Crumbs facilitates single-fiber tracing in low contrast, dense, volumetric image datasets. Scientists immerse themselves inside a data volume, using Crumbs rendering tools to view specific regions and drop markers along a structure of interest. NCSA Crumbs developers include Rachael Brady, John Pixton, George Baxter, Patrick Moran, and Clint Potter. Bridget Carragher of the Beckman Institute and Andrew Belmont from the UIUC Department of Cell and Structural Biology are also members of the Crumbs team.
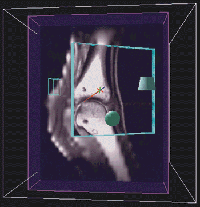
Martinelli's goal is to use 3D rendered images from MRI datasets to produce detailed anatomical descriptions of the equine metacarpophalangeal joint, especially articular cartilage, tendons and ligaments, and bone marrow. Creating anatomical descriptions of these 3D structures is difficult using traditional 2D displays of slice data or rendered volumes. Although Martinelli and other members of his group have spent considerable time trying to achieve the necessary segmentation using traditional volume analysis packages, their efforts have met with little success.
Segmentation was quickly and accurately achieved using the Crumbs environment. After Martinelli's MRI-generated fetlock data were put into the CAVE, he learned the Crumbs program and traced the cartilage in a single morning. The points marked using Crumbs will be used to generate a geometric model of the cartilage that can then be visualized in the context of the fetlock as a whole. These techniques -- segmentation with Crumbs as well as model generation -- have great potential for both research applications and for the creation of effective teaching tools.
Little is known about what happens during and just after fertilization, the time when a sperm enters the egg. Karr's basic research program uses the fruitfly as a model, but his results could be applied to mammalian or human fertilization questions. This summer Karr discovered "a whole new area of biology" while working with Crumbs.
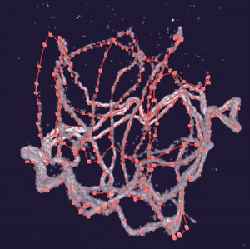
Karr developed antisperm antibodies to use with confocal microscopy to image sperm lengths in twelve species of Drosophila. Four species were traced and measured in the CAVE. The complexity of the sperm structures traced using Crumbs is apparent in the figure above. The extreme length and compaction of the sperm in the egg prevented measuring its length using a standard workstation environment. He is excited about the contributions Crumbs is making to his research.
"The CAVE lets me look at things I could not have seen before, and Crumbs lets me trace the path of the sperm inside the egg. Think about trying to follow an unraveling ball of yarn inside an egg, and you have an idea of how difficult this would be without software like Crumbs," explains Karr. "The CAVE's 3D environment was needed to trace the entire length of the sperm. We made some major strides this summer at NCSA." The ability to trace the 3D structure of supramolecular complexes like sperm inside the egg will continue to provide unprecedented new information about structure and function of complex biological systems not previously amenable to experimental examination.
Karr plans to extend these studies to mammalian fertilization and to use the CAVE to carefully compare the 3D shape of sperm in various species of Drosophila.

Another feature of the chick embryo dataset that they were able to trace was the fusion of the dorsal aorta. This vessel is a paired structure during early development, but, as development progresses, it begins fusing into a single structure midway down its length. Eurell and Sinn-Hanlon were able to trace the paired vessel to its fusion site and then back again to a paired structure as they moved from the embryo's caudal (tail) region to its head region. Long-range plans for the project include the visualization of more datasets of earlier and later embryos (24-hour-old chick and 10-mm long pig). Each dataset has features that are difficult for students to conceptualize using current teaching methods. Currently students have to imagine a 3D structure using serial sections of the embryo as viewed individually under the microscope. Seeing 3D representations of anatomical features will be a unique "field trip" for students.
In preparation for SC'95, the NCSA research team recently used a modified MRI system to test the distributed remote data acquisition interface of Crumbs. To demonstrate the capability, they put an earthworm in a soil sample and imaged it using an MRI system that sent a new volume to the Crumbs remote server every 30 seconds. The server forwarded the image to the CAVE, taking 2 to 3 seconds to update the entire data volume. The Crumbs package tracked the worm's structure, labeling its location, so changes in the worm's position were easily detected.
Further refinement of Crumbs is anticipated in 1996 as additional collaborations take place in other disciplines.
Return to the Table of Contents.